Research
I’m engaged in projects that span wild animal welfare, conservation genetics, and computational methods.
Developing indicators for wild animal welfare
All vertebrates (and some invertebrates) demonstrate sentience, the ability to experience positive and negative mental states (e.g., safety or fear). Sentience possibly evolved to improve decision making, driving individuals away from harmful stimuli and toward fitness-enhancing behaviors. Measuring welfare in wild animals has the potential to explain behavior, predict population trends, and aid in conservation efforts, but is rarely explored beyond the ecology of fear. I’m currently engaged in projects to 1) identify which indicators of welfare may be most valid and useful to measure wild animal welfare, and 2) assess whether vocalizations are a valid, non-invasive indicator of welfare in wild house sparrows. See Wild Animal Initiative for more information on this exciting and growing field.
Comparing population structure in historical and modern salmonids
Steelhead/rainbow trout (Oncorhynchus mykiss) are native to California, where most of their populations are threatened or endangered. Over the past century, California waterways and reservoirs have been stocked with domesticated hatchery forms of the species, possibly resulting in introgression. We compared whole-genome sequencing data from a set of steelhead museum specimens collected prior to hatchery stocking with contemporary samples from the same locations. Most populations showed minimal introgression, indicating cautious optimism that a century of hatchery stocking has had minimal impact on the population genetic structure of California coastal steelhead. Manuscript in preparation.
Identifying unique adaptive variants in an extinct species
Distinguishing functional from neutral variation is an important challenge in conservation genetics. We developed GWAC, a supervised machine learning framework to prioritize genome-wide variants by functional impact. We are applying GWAC to the heath hen, an extinct prairie grouse that was native to the New England coast. The last population of heath hens inhabited a rare ecosystem, the sandplain grasslands, which extant prairie grouse species do not tolerate. We are using GWAC to identify unique adaptations that allowed the heath hens to thrive in this environment. We anticipate GWAC will be used to more accurately estimate conservation metrics such as genetic load and adaptive capacity. This is a collaborative project with Revive and Restore.
Predicting the impact of structural variants on human disease
Genome sequencing has revolutionized the clinical management of rare diseases, and it often leads to diagnoses after standard methods have failed. However, at least half of cases remain undiagnosed. Structural variants (genomic alterations larger than 50 nucleotides, such as deletions and duplications) explain a portion of these undiagnosed cases. We developed a supervised learning method to determine whether a candidate structural variant is likely to cause disease.
Tracking improvements in genetic variant catalogs over time
Researchers have cataloged hundreds of thousands of genetic variants associated with disease. Clinicians use these catalogs to interpret genetic testing results, and the presence of these variants can lead to life-altering medical procedures. Yet, these databases are incomplete and contain errors. We found that misclassified variants were more prevalent in African ancestry individuals. However, databases are improving over time as population-specific allele frequency databases grow.
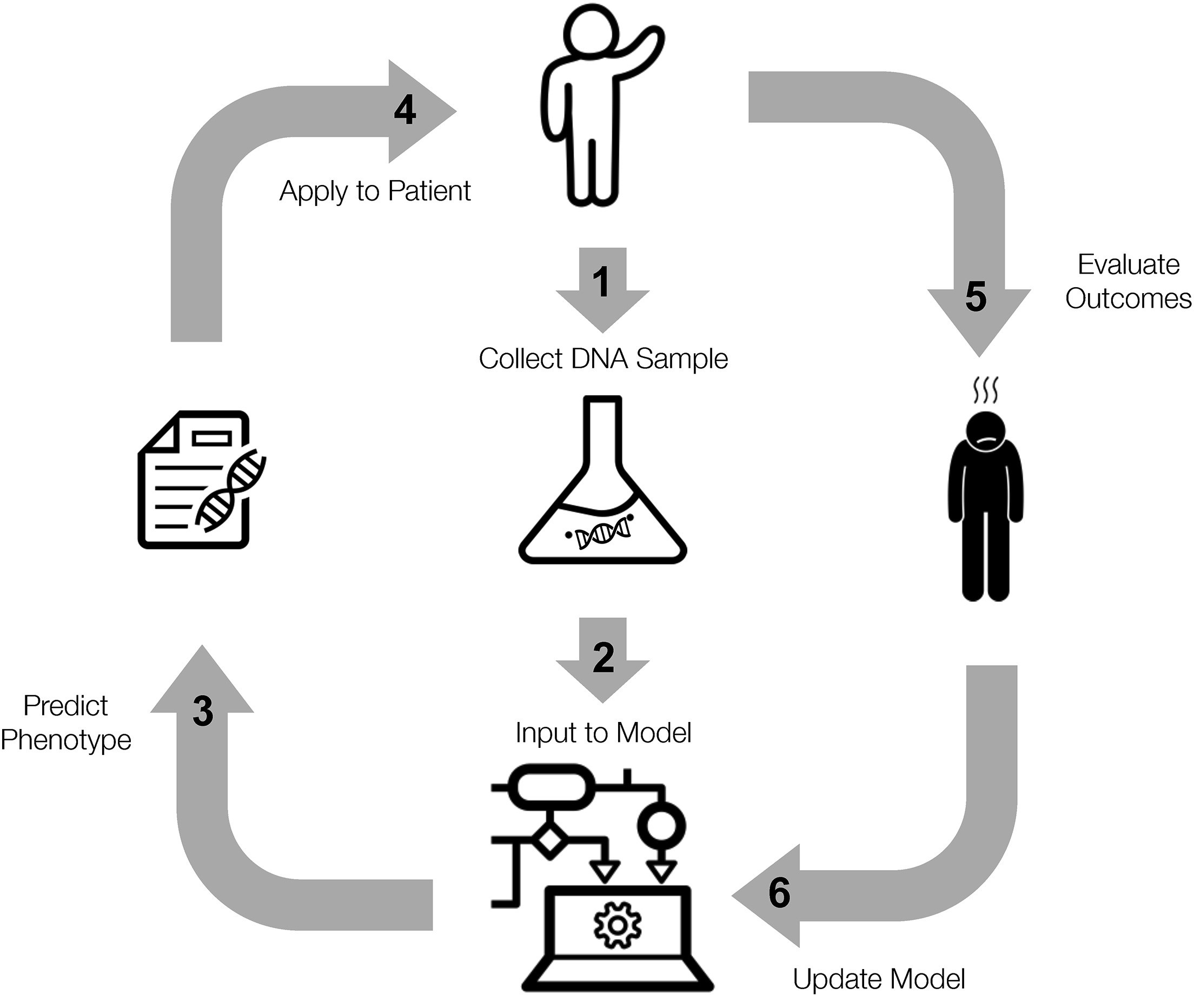
Machine learning to improve the interpretation of rare variants
Precision medicine promises to tailor treatment to the unique genetic variants in an individual. Yet the majority of missense variants in clinically important genes have uncertain clinical significance. They are also extremely rare. When these variants do appear, clinicians have little to no data to determine a likely disease course or effective treatment. To address these challenges, we propose a genomic learning healthcare system, while considering the technical and ethical challenges.